
Mosquitoes transmit the virus that causes the often-deadly disease dengue fever.
Credit: Wikimedia Commons
Download the high-resolution JPG version of the image. (1.2 MB)

Pagoda in Myanmar, site of a 2001-2002 epidemic of dengue fever.
Credit: Ruian Ke
Download the high-resolution JPG version of the image. (168 KB)
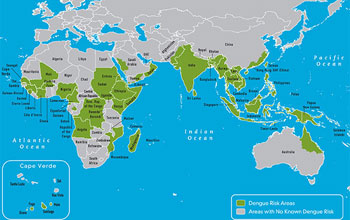
Map showing where dengue has occurred in the Eastern Hemisphere.
Credit: CDC
Download the high-resolution JPG version of the image. (81 KB)
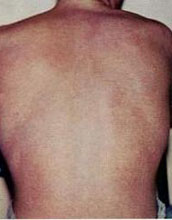
The typical rash seen in people with dengue fever.
Credit: Wikimedia Commons
Download the high-resolution JPG version of the image. (9 KB)

Dengue is also on-the-march across the Western Hemisphere.
Credit: CDC
Download the high-resolution JPG version of the image. (67 KB)

Workers in the southern U.S. in the 1920s dig a drainage ditch to control mosquitoes.
Credit: Wikimedia Commons
Download the high-resolution JPG version of the image. (82 KB)
The following is part four in a series on the NSF-NIH Ecology and
Evolution of Infectious Diseases (EEID) Program. For part one, please
see: Cool Cat in a Hot Zone. For part two: Snails in the Waters, Disease in the Villages. For part three: Underwater Whodunit: What's Killing Florida's Elkhorn Coral?.
It's
2001 in Myanmar (formerly known as Burma), a country in Southeast Asia.
Almost 200 people have died, and more than 15,000 are ill--all having
contracted dengue fever.
Dengue is a disease transmitted by
mosquitoes and caused by four types of dengue virus. Infection may not
result in symptoms, or may cause mild, flu-like illness--or hemorrhagic
fever.
Dengue virus infects some 50-100 million people annually in Southeast Asia, South America and parts of the United States.
In 1998, a pandemic of dengue resulted in 1.2 million cases of dengue hemorrhagic fever in 56 countries.
In
Myanmar, dengue is endemic. The disease has occurred there in three- to
five-year cycles since the first recorded outbreak in 1970. Each one
has been more deadly.
What caused the widespread infection in
Myanmar in 2001, a disease that resulted from one type of dengue virus,
DENV-1? For more than a decade, researchers have been working to solve
the puzzle.
All viruses not created equal
Could the DENV-1 in Myanmar have been different in some way, perhaps "defective"?
Defective
viruses result from genetic mutations or deletions that eliminate
essential functions. They're generated in viruses with high mutation
rates, but were believed to be unimportant.
But it now appears that defective viruses may be able to play a critical role in the spread of disease.
In a paper published this week in the journal PLoS Pathogens,
scientists funded by the National Science Foundation (NSF) report a
significant link between one such defective virus and the high rate of
transmission of DENV-1 in Myanmar in 2001.
"The idea has always
been that defective viruses are either meaningless or detrimental," says
James Lloyd-Smith, an ecologist and evolutionary biologist at
University of California, Los Angeles.
"We've found the
opposite--that the defective virus is actually helping the normal,
functional virus. It's bizarre and hard to believe, but the data are the
data."
"We've shown that the defective virus not only goes with
the normal virus, but increases the transmission of that virus," says
scientist Ruian Ke, also of UCLA.
While defective viruses can't
complete their life cycle on their own, if they're able to get into the
same cell with a non-defective virus, they can "hitch-hike" with the
non-defective one and propagate.
Deadly outbreak of DENV-1
The
research team--James Lloyd-Smith; Ruian Ke; John Aaskov, a virologist
at Queensland University of Technology in Brisbane, Australia; and
Edward Holmes, a biologist at the University of Sydney--found that the
presence of a defective DENV-1 virus may have led to a spike in dengue
fever cases in Myanmar during 2001-2002.
"The causes of epidemics
are much more complicated than we thought," says Sam Scheiner, NSF
program director for the joint NSF-National Institutes of Health Ecology
and Evolution of Infectious Diseases (EEID) Program. At NSF, EEID is
funded by the Directorates for Biological Sciences and Geosciences.
In addition to EEID, the research was supported by NSF's Advancing Theory in Biology Program.
"Pathogens
can depend on the presence of other microbial species or, as in this
case, other varieties of the same species," says Scheiner.
"Understanding these interactions is critical for predicting when the
next epidemic might occur--and how to prevent it."
In the study, Ke designed a mathematical model to learn how the defective DENV-1 virus interacted with the normal virus.
Aaskov
and Holmes collected genetic sequences from the defective viruses from
15 people sampled over an 18-month period in Myanmar. All were infected
with DENV-1 virus; nine were also infected with the defective version.
Ke
discovered that the lineage of defective viruses emerged between June
1998 and February 2001; it spread through the population until at least
2002.
The following year, the lineage appeared in the South Pacific island of New Caledonia, carried there by a mosquito or a person.
The
scientists analyzed the genetic sequences of the defective and normal
viruses to estimate how long the defective virus had been transmitting
in the human population.
"We can see from the gene sequence of the
defective version that it's the same lineage, and is a continued
propagation of the virus," says Lloyd-Smith.
"From 2001 to 2002,
it went from being quite rare to being in all nine people we sampled
that year," says Lloyd-Smith. "Everyone sampled who was getting dengue
fever was getting the defective version along with the functional virus.
"It rose from being rare to being very common in just one year."
Most
surprisingly, say the scientists, the combination of the defective
virus with the normal virus was "more fit" than the normal dengue virus
alone.
"What we've shown is that this defective virus, which
everyone had thought was useless or even detrimental to the fitness of
the functional virus, actually appears to have made it better able to
spread," Lloyd-Smith says.
Ke calculated that the defective virus
makes it at least 10 percent more transmissible. "It was spreading
better with its defective cousin tagging along than on its own," says
Lloyd-Smith.
It takes two (viruses) to tango
The functional virus and defective virus travel in unison. The two transmit together in an unbroken chain.
"That's
not just a matter of getting into the same human or the same
mosquito--they need to get into the same cell inside that human or
mosquito in order to share their genes, and for the defective version to
continue hitchhiking," says Lloyd-Smith.
"We're gaining insights
into the cellular biology of how dengue is infecting hosts. It must be
the case that frequently there are multiple infections of single cells."
The defective virus appeared one to three years before the major epidemics in 2001 and 2002.
"One
could imagine that if you build an understanding of this mechanism, you
could measure it, see it coming and potentially get ahead of it," says
Lloyd-Smith.
Defective viruses: disease transmitters beyond dengue?
Might defective viruses play a role in the transmission of the flu, measles and other diseases?
"There are a few signs that this phenomenon may be happening in other viruses," Lloyd-Smith says.
"We
may be cracking open the book on the possible interactions between
normal, functional viruses and the defective ones that people thought
were just dead-ends.
"These supposedly meaningless viruses may be having a positive effect--positive for the virus, not for us.
"There's
great variation from year to year in dengue epidemics in various
locations, but we don't understand why. This is a possible mechanism."
Why would a defective virus increase transmission of a disease?
Lloyd-Smith offers two hypotheses.
One
is that the presence of the defective virus with the functional virus
in the same cell makes the functional virus replicate better within the
cell by an unknown mechanism.
"It might give the virus flexibility
in how it expresses its genes, and may make it more fit and better able
to reproduce under some circumstances," Lloyd-Smith says.
A second idea is that the defective virus may be interfering with the disease-causing virus, making the disease less intense.
People
then have a milder infection, and because they don't feel as sick,
they're more likely to go out of their homes and spread the disease.
In
conducting the research, Lloyd-Smith and Ke combined genetic sequence
analyses with sophisticated mathematical models and bioinformatics.
"We
were able to show that this defective virus transmitted in an unbroken
chain across this population in Myanmar for a year-and-a-half,"
Lloyd-Smith says.
"Without gene sequencing, we wouldn't have been able to establish that."
The
biologists hope their work will help turn the tide of the next deadly
outbreak of dengue in Myanmar--and in other tropical countries around
the globe.
| -- | Cheryl Dybas, NSF (703) 292-7734 cdybas@nsf.gov |
Related Websites
NSF Special Report: Ecology and Evolution of Infectious Diseases:
NSF Special Report: Ecology and Evolution of Infectious Diseases:
http://www.nsf.gov/news/special_reports/ecoinf/index.jsp
PLOS Pathogens journal paper:
PLOS Pathogens journal paper:
http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1003193
UCLA: defective dengue viruses:
UCLA: defective dengue viruses:
http://newsroom.ucla.edu/portal/ucla/defective-virus-surprisingly-243742.aspx
U.S. Centers for Disease Control and Prevention: Dengue:
U.S. Centers for Disease Control and Prevention: Dengue:
The National Science Foundation (NSF)
Guillermo Gonzalo Sánchez Achutegui
ayabaca@gmail.com
ayabaca@hotmail.com
ayabaca@yahoo.com
Inscríbete en el Foro del blog y participa : A Vuelo De Un Quinde - El Foro!

No hay comentarios:
Publicar un comentario